Research
In Vitro
Actin and Microtubule Networks Geometry
Actin networks
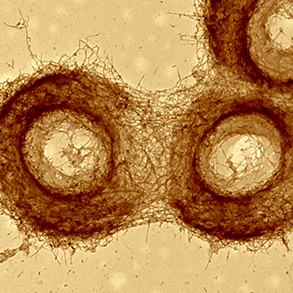
Actin is an asymmetric protein that can self-assemble to form polarized actin filaments. Actin filaments can interact to form actin networks. Actin networks can self-organize into two main types of structures in cells: bundles (or fibers) made of aligned long filaments and meshworks made of branched and intermingled short filaments. Our aim is to reconstitute both and investigate : 1- how the spatial distribution of actin nucleators directs the growth and assembly of filaments to form defined network geometries in 2D and 3D, 2- how the contraction and disassembly processes regulate networks turn-over and permanent renewal. In both cases we try to relate network dynamics to its physical properties and the eventual production of mechanical forces. A particular attention is devoted to the study of the role of actin network geometry and size.





Microtubule networks
Similar to the formation of actin filaments from the self-assembly of actin monomers, tubulin forms asymmetric dimers that can self-assemble into microtubules. Compared with actin filaments, microtubules are much more rigid, and almost straight in the dimensions of a single cell. Microtubules can sustain much higher compression forces than actin filament but are not as numerous as actin filaments. In most animal cells, the MT network forms as an aster in which microtubules radiate from the microtubule organizing center (MTOC). As cells divide, the MTOC is duplicated and the network forms a bipolar spindle. Our aim is to investigate the role of microtubule mechanics and microtubule-associated proteins in the establishment of the various microtubule network architectures during interphase and mitosis.


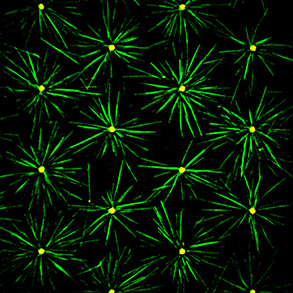
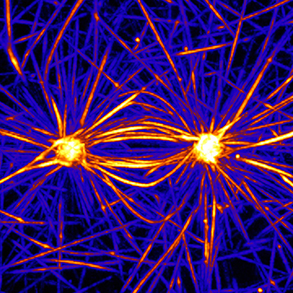
Actin-microtubule crosstalk
Actin and microtubule networks interact by many ways including common signaling pathways, specific crosslinkers or via less specific steric interactions. We currently work on the reconstitution of such interactions and their contribution to the establishment of coherent heterogeneous cytoskeleton networks architectures.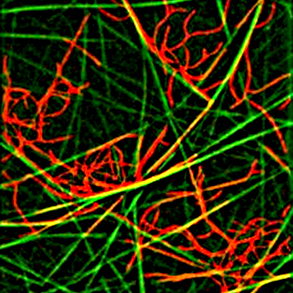
Budding Yeast as a model system
The efficiency of DNA strand homologous recombination in budding yeast makes it an organism of choice to remove one or several genes of interest. In addition, yeast culture methods offer the possibility to extract cell cytoplasmic content and work with large amounts of material. We use yeast extracts to investigate cell cytoskeleton self-organization in complex, because made of whole cellular content, but controlled, because genetically modulated, biochemical conditions. Adding more and more compounds to mixtures of purified proteins and removing proteins from cell extracts will allow us to bridge in vitro and in vivo approaches and benefit from a broad range of biochemical conditions to investigate cytoskeleton self-organization laws.