Directed Cytoskeleton Self-Organization
From molecules to cell polarity
The reproducible shape and spatial organization of organs imply the existence of physical rules directing the assembly of complex biological structures. Organ shape and function depend on cell architecture and polarity, which are both supported by cell cytoskeleton networks. The formation of controlled and reproducible geometrical structures relies on the self-organization properties of these networks. Our aim is to unravel the physical processes underlying cytoskeleton self-organization processes and to formulate the rules directing their spatial organization.
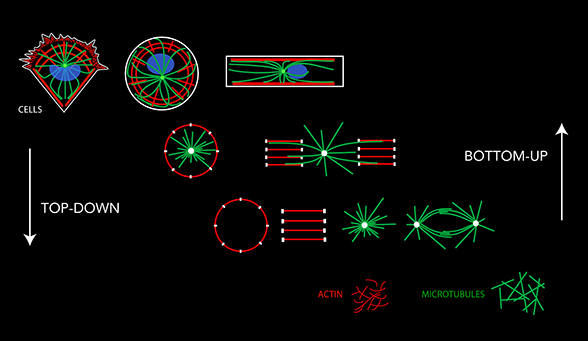
Actin filaments and microtubules form such complex intricate networks in cells that it is difficult to identify the principles of their self-organization. Our rationale is that these principles can manifest themselves in a reproducible, and therefore understandable, way only in response to defined geometrical cues. Thus, to study the geometrical and mechanical rules underlying cytoskeleton self-organization we used microfabrication tools in order to control and manipulate the spatial boundary conditions the cytoskeleton networks are sensitive to.These tools allow us to analyze and quantify actin and microtubule networks in cells of controled and regular shapes. Considering that the complexity of the intra-cellular biochemical conditions may partially hinder the physical rules we want to investigate, we are also developing alternative methods to analyze cytoskeleton self-organization in controlled biochemical conditions in vitro by mixing, in defined proportions, the individual cytoskeleton components. Cells extracts and cytoplastesallow us to bridge the gap between these two approaches. The simplification of the cellular approach, the top-down way, and the complexification of the biochemical approach, the bottom-up way, should eventually encounter and provide us a continuous experimental platform to analyze the physics of cytoskeleton networks and morphogenesis from molecules to cells.
